What would happen if a Pakistani consumer thought she was buying chamomile to help her sleep or hyssop to help with a cough, but instead the herbs were misidentified?
“In Pakistan, herbal products are a primary source of medicines, yet quality control for raw plant material sold to individuals or companies is very limited,” says Dr. Allan Showalter, Professor of Environmental and Plant Biology in a conference abstract. “Large-scale cultivation of medicinal plants is not prevalent, so the bulk of raw material is imported from other countries or collected from the wild, with no provenance. Roots, bark, twigs, leaves, flowers, and seeds are sold under common names in local languages, so the potential for misidentifications and mixed collections is high.
“Misidentifications or adulteration of authenticated materials can lead to reduced effectiveness of herbal products or accidental poisonings. Barcoding provides a way to confirm the identification of raw plant material and establish a level of quality assurance. We have generated barcodes from 43 medicinal plant species, representing 23 different families, using the rbcL, matK, and psbA-trnH spacer regions. Six market samples have been documented as adulterated or entirely misidentified, including Matricaria recutita var. chamomilla (chamomile) and Hyssopus officinalis (hyssop). Comparisons of medicinal plant barcodes with those of sister species have indicated that the psbA-trnH spacer is most useful for distinguishing species, followed by matK and rbcL,” he concludes.
Showalter and Dr. Melanie Schori ’10PhD, a postdoctoral fellow and College of Arts & Sciences alum, are traveling to Kunming, China, to present their research at the fifth annual Barcode of Life Conference in October.
Showalter will give an oral presentation on “DNA Barcoding Medicinal Plants from Pakistan.” See Sept. 27 colloquium: Zabta Shinwari, Quaid-i-Azam University, “Molecular Systematics and Applied Ethnobotony in Pakistan.”
Schori will present on “Improved PCR and Barcoding Data Analysis.” She acknowledges several other people at Ohio University who have contributed to the research she will present:
- AlexaRae Kitko, an Honors Tutorial College student
- Dr. Wei Lin, Associate Professor of Mathematics
- Yichao Li, a Ph.D. student in the Bioinformatics program
- Dr. Lonnie Welch, Stuckey Professor of Electrical Engineering and Computer Science at Ohio University
Lin, Li, and Welch have developed a computer program, still in the testing phase, that is the feature of the second part of her talk.
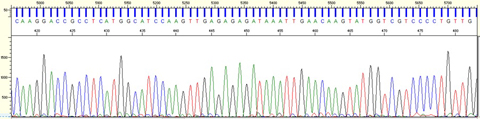
The image below is part of a DNA sequence for dill. “This is what the sequence looks like when we get it back from the Genomics Facility. The different colored peaks correspond to the four nucleotides that make up DNA. The photo above is one of Allan’s photos from our first trip to Pakistan,” said Schori. “It is a shop where people can purchase medicinal herbs, most of which are in bins, drawers, or bags. It’s very different from how we buy herbal products in the United States.”
“DNA barcoding of medicinal plants is an effective way to identify adulterated or contaminated market materials, but it can be quite challenging, both in generating barcodes and analyzing the data to determine discrimination power,” says Schori in her abstract. “Successful PCR of barcoding regions is often inhibited by desirable secondary metabolites in the medicinal plants. However, modifications of extraction methods, primer sequences, and the use of an engineered polymerase can usually overcome PCR problems.
“Sequence data were obtained for medicinal plants from 24 different families at rates of 96% for rbcL and 79% for matK. Data were then compared to sister species sequences in GenBank to assess which gene region was most informative for the medicinal species of interest. Unfortunately, many sequences in GenBank contain errors. An algorithm was developed to ‘clean’ alignments and identify problematic sequences, facilitating their removal or correction. A second algorithm can then determine how many positions can be used to perfectly discriminate between pairs of species, thus highlighting which gene region is most useful for identifying species within a particular genus.”






















Pingback: Medicinal herbs Info | Medicinal herbs